Peptide libraries are powerful tools for bioactive molecule discovery and development, and custom peptide library manufacturing is a specialised service that provides the highest quality peptides for your research needs.
We offer a wide range of peptide library designs, including linear peptides, cyclic peptides, and peptide mixtures.
Genosphere Biotechnologies offer custom peptide libraries synthesis in 96-wells microplate formats. Whether you intend to screen peptides for enzyme substrate profiling, elucidate substrate specificities, protein-protein interaction region mapping, or antibody epitope mapping, we provide cost-effective fmoc-based simultaneous multi-peptide synthesis technologies to fit your needs.
With that in mind our team has developed two service packages:
(i) Specification: Basic screening-grade peptide library
For economical initial or large scale peptide screens.
Peptide length : 5-20 aa
Free C- & N- termini
Amount: 1 to 2 mg
Purity: Screening-grade, desalted synthesis (average purity for 12mer >75%)
Analysis: Mass spectrum for each peptide
Minimum order: 48 peptides
Leadtime: 1 plate, average 2 weeks
(ii) Specification: Pure qualification-grade peptide library
For exploration studies and validation of identified target sequences.
Peptide length : 5-20 aa
Amount: 1 to 2 mg
Modifications: N-C-capping, biotin, flurorophores, unnatural amino acids
Purity choice: >80% ; >90% ; >95%
Analysis: RP-HPLC and Mass spectrum for each peptide
Minimum order: 24 peptides
Leadtime: average 3 weeks
Genosphere Biotechnologies’ peptide libraries are routinely used to screen for antigenic peptides, receptor ligands, antimicrobial compounds, and enzyme inhibitors identification. Some of the applications include:
- Epitope mapping studies
- Vaccine research
- High-throughput protein-protein interaction analysis
- Kinase assays
Our experienced team of chemists and engineerings will work closely with you to develop a custom peptide library that meets your exact specifications. We use the latest technology and techniques to ensure that our peptide libraries are of the highest quality and purity. Furthermore, our peptide libraries are rigorously tested for accuracy and consistency.
With our custom peptide library manufacturing services, you can be sure that your research project will be successful. Our team is committed to providing the best service and highest quality products to our customers, so you can trust that your peptide library will be of the highest standard.
Synthetic peptide library design
Many synthetic library design have been successfully developed for various applications.
Mutational peptide library
Mutational peptide library design involves The synthesis of peptides with single or multiple point mutations. This approach is useful for understanding the effect of mutations on the activity of a peptide, as well as for designing peptides with desired activities.
Truncation peptide library
Truncation peptide library design involves synthesising peptides with truncated sequences. This approach is used to identify shorter peptides with the same activity as longer peptides. It is also useful in determining the minimum sequence required to produce a certain activity, or the minimum size of a peptide necessary for a certain function.
Cyclic peptide library
Cyclic peptide library design involves synthesising peptides that are cyclic in structure. This approach is useful for understanding the role of the cyclic structure of a peptide in its activity, as well as for designing peptides with desired activities.
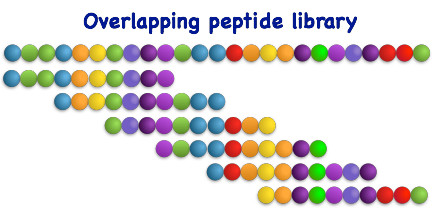
Overlapping peptide library
Overlapping peptide libraries are constructed by breaking down a protein or peptide sequence into multiple short peptides that overlap with each other. The length and offset of these peptides are determined in advance and are typically set to be 8 to 20 amino acids in length, with an offset of 2-4 amino acids.
This approach provides comprehensive coverage of the target protein and enables the identification of specific peptides that bind to the target molecule through techniques such as mass spectrometry or protein-protein interaction analysis. The optimal length and offset for epitope mapping have been determined to be within the aforementioned range.
In vitro evolution peptide library
In vitro evolution library design involves creating a library of peptides by subjecting them to in vitro selection and amplification. This method is useful for discovering new peptides with desired activities, or for engineering existing peptides to have specific activities. The library is created by subjecting peptides to selection criteria, such as binding to a target molecule, and then amplifying the successful peptides to generate a large library. This approach is often used in combination with other methods, such as random library design and recombinant library design.
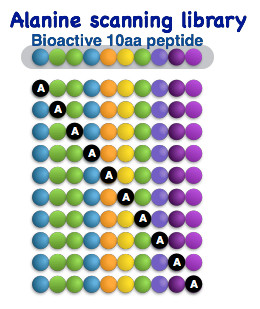
Alanine scanning peptide library
Alanine scanning peptide library design involves replacing a single amino acid residue in a peptide with alanine, and then synthesising a library of peptides where each one contains a different alanine substitution. This approach is useful for determining which amino acid residues are involved in the activity of a peptide, as well as what the relative importance of each residue is.
Positional scanning peptide library
Positional Scanning Library is a key method for peptide sequence optimization. By sequentially replacing selected amino acid residues with all other natural amino acids, it has the ability to identify potentially more favorable residues at specified positions for improved peptide activity.