Las bibliotecas de péptidos son potentes herramientas para el descubrimiento y desarrollo de moléculas bioactivas. La fabricación de bibliotecas peptídicas a medida es un servicio especializado que proporciona péptidos de la máxima calidad para sus necesidades de investigación. Ofrecemos una amplia gama de bibliotecas de péptidos, incluyendo péptidos lineales, péptidos cíclicos y mezclas de péptidos.
Proponemos la síntesis de bibliotecas de péptidos sobre microplacas de 96 pozos. Nuestra estrategia basada en la química Fmoc de síntesis paralela nos permite ofrecerle tarifas ventajosas. Nuestras bibliotecas están perfectamente adecuadas para aplicaciones de muestreo de especificidad y de substrato enzimáticos, de identificación de regiones de interacción proteína- proteína y también de cartografía de sitios de reconocimiento de anticuerpos.
Teniendo esto en cuenta, nuestro equipo ha establecido dos tipos de servicio:
(i) Especificación: Biblioteca básica de péptidos para cribado
Para el cribado inicial o a gran escala de péptidos a bajo coste.
Longitud del péptido: 5-20 aa
Terminales C y N libres
Cantidad: 1 a 2 mg
Pureza: Síntesis desalada, calidad de cribado (pureza media para 12mer >75%)
Análisis: espectro de masas para cada péptido
Pedido mínimo: 48 péptidos
Plazos: 1 placa, 2 semanas de media
(ii) Especificación: Biblioteca de péptidos puros para validación/calificación
Para estudios de exploración y validación de secuencias diana identificadas.
Longitud del péptido: 5-20 aa
Cantidad: 1 a 2 mg
Modificaciones: N-C-capping, biotina, fluroróforos, aminoácidos no naturales
Elección de la pureza: >80%; >90%; >95%.
Análisis: RP-HPLC y espectro de masas para cada péptido
Pedido mínimo: 24 péptidos
Plazo de entrega: 3 semanas de media
Las bibliotecas de péptidos de Genosphere Biotechnologies se utilizan habitualmente en proyectos que implican el cribado de péptidos antigénicos, ligandos de receptores, compuestos peptídicos antimicrobianos y la identificación de inhibidores enzimáticos. Algunas de estas aplicaciones son
- Mapeo de epítopos
- Investigación y desarrollo de vacunas
- Análisis de alto rendimiento de interacciones proteína-proteína
- Estudios de actividad cinasa
Nuestro experimentado equipo de químicos y bioingenieros trabaja en estrecha colaboración con usted para desarrollar una biblioteca de péptidos personalizada que cumpla exactamente sus especificaciones. Utilizamos la tecnología más avanzada para garantizar que nuestras bibliotecas de péptidos sean de la máxima calidad y pureza. Además, nuestras bibliotecas peptídicas se someten a rigurosas pruebas de validez y calidad.
Con nuestro servicio de fabricación de bibliotecas peptídicas personalizadas, puede estar seguro de que su proyecto de investigación será un éxito. Nuestro equipo se compromete a proporcionar el mejor servicio y los productos de mayor calidad a nuestros clientes. Así que puede estar seguro de que su biblioteca de péptidos será de la más alta calidad.
Diseño de una biblioteca de péptidos sintéticos
Se han desarrollado con éxito numerosas bibliotecas sintéticas para diversas aplicaciones.
Biblioteca peptídica mutacional
El diseño de una biblioteca peptídica mutacional implica la síntesis de péptidos con mutaciones puntuales únicas o múltiples. Este enfoque es útil para comprender el efecto de las mutaciones en la actividad de un péptido, así como para diseñar péptidos con las actividades deseadas.
Biblioteca de péptidos truncados
El diseño de una biblioteca de péptidos truncados consiste en sintetizar péptidos con secuencias truncadas. Este enfoque se utiliza para identificar péptidos más cortos con la misma actividad que los péptidos más largos. También es útil para determinar la secuencia mínima necesaria para producir una determinada actividad, o el tamaño mínimo de un péptido necesario para una determinada función.
Biblioteca de péptidos cíclicos
El diseño de una biblioteca de péptidos cíclicos consiste en sintetizar péptidos con una estructura cíclica. Este enfoque es útil para comprender el papel de la estructura cíclica de un péptido en su actividad, así como para diseñar péptidos con las actividades deseadas.
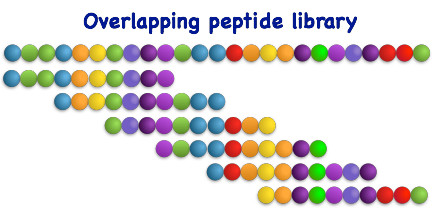
Biblioteca de péptidos solapados
Las bibliotecas de péptidos solapados se construyen dividiendo una proteína o secuencia peptídica en varios péptidos cortos solapados. La longitud y el desplazamiento de estos péptidos se determinan de antemano y suelen ser de entre 8 y 20 aminoácidos, con un desplazamiento de 2 a 4 aminoácidos.
Este enfoque proporciona una cobertura completa de la proteína diana y permite la identificación de péptidos específicos que se unen a la molécula diana mediante técnicas como la espectrometría de masas o el análisis de interacción proteína-proteína. Se determinó que la longitud y el desplazamiento óptimos para la cartografía de epítopos se encontraban dentro del intervalo mencionado.
Biblioteca de péptidos de evolución in vitro
El diseño de bibliotecas de evolución in vitro consiste en crear una biblioteca de péptidos sometiéndolos a selección y amplificación in vitro. Este método es útil para descubrir nuevos péptidos con las actividades deseadas o para modificar péptidos existentes de modo que tengan actividades específicas. La biblioteca se crea sometiendo los péptidos a criterios de selección, como la unión a una molécula diana, y amplificando después los péptidos seleccionados para generar una gran biblioteca. Este enfoque se utiliza a menudo en combinación con otros métodos, como el diseño aleatorio de bibliotecas y el diseño de bibliotecas recombinantes.
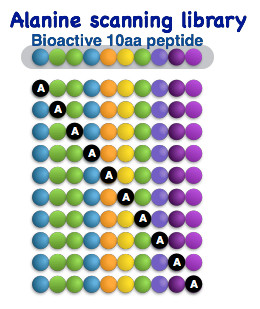
Biblioteca de péptidos de barrido de alanina
El diseño de una biblioteca de péptidos de escaneo de alanina implica la sustitución de un único residuo de aminoácido en un péptido por alanina y, a continuación, la síntesis de una biblioteca de péptidos, cada uno de los cuales contiene una sustitución de alanina diferente. Este enfoque es útil para determinar qué residuos de aminoácidos intervienen en la actividad de un péptido, así como la importancia relativa de cada residuo.
Biblioteca peptídica de barrido posicional
La biblioteca de exploración posicional es un método clave para optimizar las secuencias peptídicas. Al sustituir secuencialmente residuos de aminoácidos seleccionados por todos los demás aminoácidos naturales, identifica residuos potencialmente más favorables en posiciones específicas para mejorar la actividad del péptido.